CRISPR human sgRNA libraries
CRISPR human sgRNA libraries
LabOmics offers GeneCopoeia's CRISPR sgRNA libraries for high-throughput knockout of human genes in specific or custom gene groups or pathways. Loss-of-function screening by gene knockout is a powerful tool for systematic genetic analysis in mammalian cells, facilitating gene discovery, genome-scale functional interrogation (e.g. signal transduction pathways) and drug discovery (e.g. target identification and drug mechanism studies).
The Genome-CRISP™ human single guide RNA (sgRNA) libraries are cloned into lentiviral vectors for dual-use (transfection or transduction) delivery methods designed for large-scale functional screens. For each targeted gene, a minimum of 2 barcoded sgRNAs targeting different regions are created, optimized and sequence-verified to ensure efficient gene knockout. Further, each sgRNA-expressing plasmid is individually cultured in E. coli before pooling, providing the best possible representation of each sgRNA in the pools. The libraries can be ordered as pools of sgRNAs in pre-defined gene families or as custom sets, and are available as lentiviral particles, transfection-ready plasmid DNA or bacterial stocks. They can be transfected or transduced into cell lines, including those stably expressing Cas9 nuclease.
Advantages
- sgRNAs are individually constructed, sequence-verified, and cultured in E. coli before pooling, ensuring high quality and comprehensive sgRNA representation.
- Premade or custom-made sgRNA libraries available
- 2 or more sgRNAs per target gene
- Flexible delivery formats such as pooled lentiviral particles, transfection-ready plasmid DNA, or bacterial bacterial stocks

Figure 1. Illustration of large scale screening with sgRNA library
Applications
- High-throughput knockout screening with many sgRNAs, either individually or in pools.
- Drug target discovery (Figure 2).
- Drug target validation.
- Phenotypic screens.
- Reporter assays.
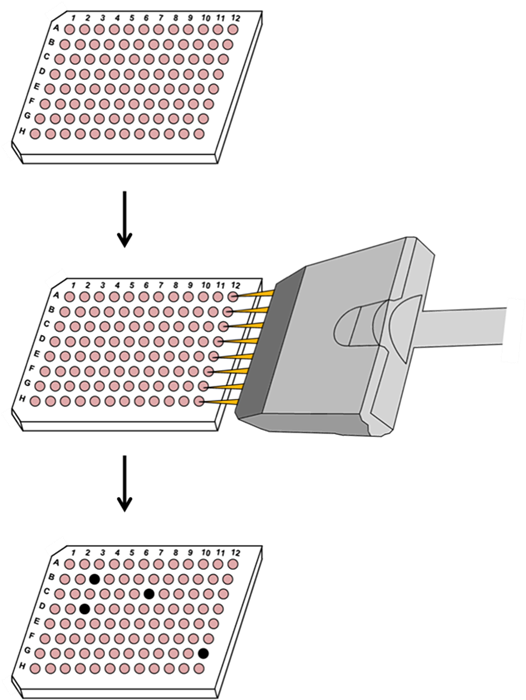
96-well plate with Cas9-expressing cells
Transduce or transfect with CRISPR library sgRNAs
Screen plates for response (e.g. cell death resulting from knockout)
Figure 2. Example application for use of the Genome-CRISP™ sgRNA libraries in drug target discovery. Top: Each well of a 96-well plate is cultured with cells stably expressing the CRISPR Cas9 nuclease.
Middle: Libraries of Genome-CRISP™ sgRNAs, either individually or in pools, are used to transduce or transfect cells by addition with a multichannel pipettor.
Bottom: Plates are screened for cell death (black wells). Each sgRNA is barcoded, so that individual or pooled sgRNAs can be identified as candidates for drug targets.
The Genome-CRISP™ CRISPR human sgRNA libraries consist of sequence-verified sgRNA target sites cloned into a lentiviral vector. The clones do not carry the Cas9 nuclease. You will need to either purchase a lentiviral Cas9 clone or a cell line stably expressing Cas9. For lentiviral transduction, we recommend using a cell line stably expressing Cas9 before transduction with the libraries.
Human sgRNA libraries
| Catalog number | Library name | Number of sgRNAs | Number of genes | Number of tubes |
|---|
| L01-LS03-F1 |
Innate kinases & ubiquitin ligases |
475 |
239 |
4 tubes, 118-119 sgRNAs each |
| L02-LS03-F1 |
Nuclear hormone receptors |
236 |
118 |
2 tubes, 118 sgRNAs each |
| L03-LS03-F1 |
Tumor metastasis genes |
114 |
57 |
2 tubes, 57 sgRNAs each |
| L04-LS03-F1 |
Oncogenes |
576 |
288 |
4 tubes, 144 sgRNAs each |
| L05-LS03-F1 |
Tumor suppressor genes |
462 |
231 |
4 tubes, 115-116 sgRNAs each |
| L06-LS03-F1 |
Protein kinases |
1316 |
658 |
10 tubes, 131-132 sgRNAs each |
| L07-LS03-F1 |
Key genes in 50 pathways |
278 |
139 |
2 tubes, 139 sgRNAs each |
 Human cell line H1299 stably expressing CRISPR Cas9, single clone1 420,00 €Human cell line H1299 stably expressing CRISPR Cas9, single clone Learn More
Human cell line H1299 stably expressing CRISPR Cas9, single clone1 420,00 €Human cell line H1299 stably expressing CRISPR Cas9, single clone Learn More Human cell line HEK293T stably expressing CRISPR Cas9, single clone1 420,00 €Human cell line HEK293T stably expressing CRISPR Cas9, single clone Learn More
Human cell line HEK293T stably expressing CRISPR Cas9, single clone1 420,00 €Human cell line HEK293T stably expressing CRISPR Cas9, single clone Learn More Human cell line HeLa stably expressing CRISPR Cas9, single clone1 420,00 €Human cell line HeLa stably expressing CRISPR Cas9, single clone Learn More
Human cell line HeLa stably expressing CRISPR Cas9, single clone1 420,00 €Human cell line HeLa stably expressing CRISPR Cas9, single clone Learn More Human cell line A549 stably expressing CRISPR Cas9, single clone1 420,00 €Human cell line A549 stably expressing CRISPR Cas9, single clone Learn More
Human cell line A549 stably expressing CRISPR Cas9, single clone1 420,00 €Human cell line A549 stably expressing CRISPR Cas9, single clone Learn More

